Olivier's paper evaluating alternative protein representations for predicting shared paralog functions
 Olivier’s paper
Olivier’s paper
Congrats to Olivier on his first publication with the lab, Evaluating Sequence and Structural Similarity Metrics for Predicting Shared Paralog Functions, up now on biorXiv.
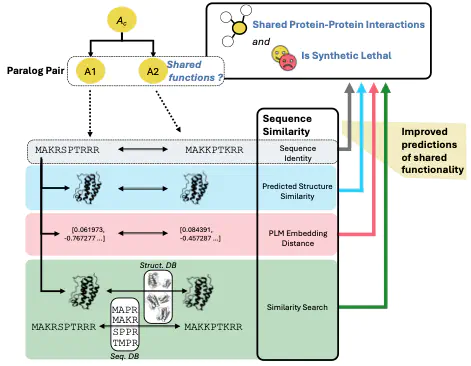
Comparisons between protein sequences are the foundation of much of modern biology, allowing us to infer evolutionary relationships between genes and assign functions to homologous proteins. These sequence comparisons are not just made across species, for inferring functions of orthologs, but also within species, for paralogs (our favourites!). For decades, protein sequence identity has been used as a proxy for the functional similarity of paralogs. However, the advent of alternative AI-derived protein representations (predicted structures from AlphaFold or embeddings from protein language models) raises the possibility that similarity metrics derived from these alternative representations might capture functional relationships that sequence identity misses. Here we set out to test this systematically – Can similarity between AlphaFold predicted structures better identify paralogs with shared functions than simple sequence identity? What about protein language model embeddings?
We evaluated two definitions of shared paralog functions (shared protein-protein interaction partners, synthetic lethality) across two species (budding yeast and human). We found that, at least for some tasks, similarity measures derived from alternative protein representations can outperform sequence identity alone (e.g. similarity of predicted AlphaFold structures better predicts shared protein-protein interactions in yeast). However, more importantly, these measures were not redundant with sequence identity, i.e. combining them with sequence identity led to improved predictions across all tasks. We show that the complementary nature of these alternative metrics applies to other definitions of shared functionality for paralogs (shared gene ontology terms). Overall it is clear that similarity metrics from these alternative protein representations can capture aspects of shared paralog functionality that are not captured by sequence identity alone.
The shared functions of paralogs are relatively easy to assess (e.g. through double perturbation experiments) and make them ideal candidates for the sort of systematic evaluation we have performed here. Looking beyond paralogs, our results suggest that combining these alternative protein representations—sequence, structure, and language model embeddings—could be highly valuable for inferring common functions in a more general sense, including for orthologs.
You can read the full paper here.
We make the full set of similarity metrics available for all yeast and human paralog pairs here. We are happy to generate them for other species too – if you are interested, send us a mail.