Anjan's paper on proteomic compensation between paralogs in cancer
 Anjan’s paralog paper
Anjan’s paralog paper
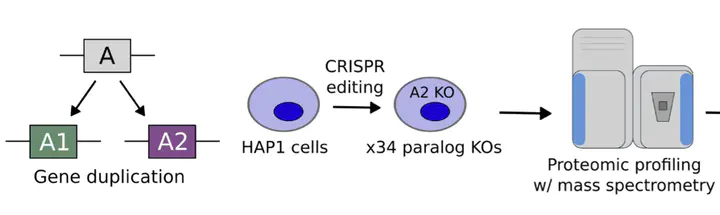
Compensation between duplicate genes (paralogs) allows cancer cells to tolerate genetic mutations that might otherwise be lethal. We wanted to know what this compensation looked at a molecular level – what are the proteins involved doing? How come losing functional genes doesn’t just just break the protein-protein interaction networks operating inside cancer cells?
We used proteomic profiling of isogenic knockouts (in collaboration with Alexander Von Kriegsheim) and large-scale analysis of tumour proteomes to explore this question systematically. We found tons of examples of proteomic compensation, where loss of one gene was associated with increased abundance of its paralog, as well as collateral loss, where loss of one gene was associated with decreased abundance of its paralog. Proteomic compensation pairs were more central in the protein interaction network, more evolutionary conserved, and more likely to be synthetic lethal.
Overall our results support a model where loss of an important gene in cancer (one that has lots of interactions, is involved in an essential function) triggers an increase in the protein abundance of its paralog. Tumour cells then become dependent on the upregulated paralog for survival.
You can read the full paper here.
You can read Anjan’s tweetorial about the paper below:
Very pleased to see my PhD work out as a preprint in bioRxiv! Here, we set out to answer a basic question— how come gene loss in cancer doesn't break protein-protein interaction networks? (1/17) pic.twitter.com/OAqK1b6BMh
— Anjan Venkatesh (@AnjanVenkatesh) October 3, 2024